Research Highlights
Advanced bioengineering for advanced biophysics
Our strong focus on the mechanistic understanding of IDPs using single molecule and super-resolution tools is both driven and driving novel tool developments for “in-cell/in situ structural biology”. This comprises of a synergistic effort of chemical/synthetic biology and precision fluorescence-based technology/nanoscopy/microfluidics development (see figure 1 left).
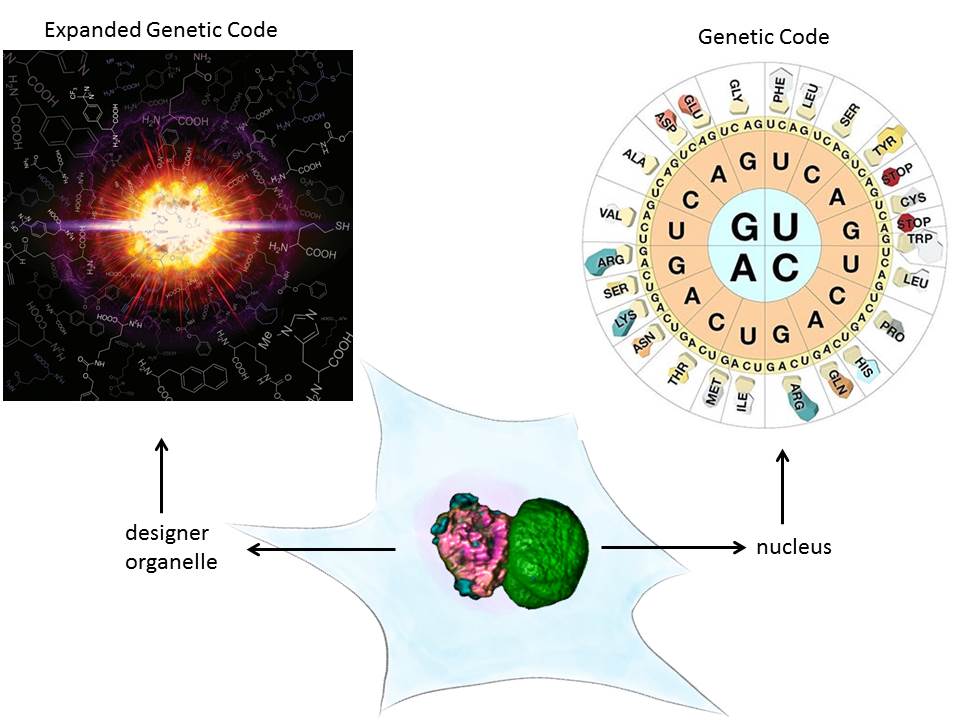
A major technical breakthrough of my lab was the ability to engineer “click”able functionalities into any protein both in vitro and in vivo. This genetic code expansion (GCE) approach has the potential to become a true GFP (fusion protein) surrogate strategy, with the major advantages being that superior synthetic dyes can be coupled with residue-specific precision anywhere in a protein. This opens up new avenues in single-molecule fluorescence and super-resolution microscopy.
We have recently also taken protein engineering to the next level, that is we have given this its own organelle. Eukaryotes use organelles to carry out specific function. We used phase separation and spatial targeting to create a membraneless organelle inside the living eukaryotic cell that carries out selectively GCE. This does not just equip the cell with a second, expanded genetic code, but also creates a factory to make noncanonical proteins without “messing” with host functionality. It also shows how easy new cellular function can be encoded by phase separation mechanisms. Membraneless organelle engineering depicts a new way to do both, study the cell in a minimally invasive way and to create new functionality.
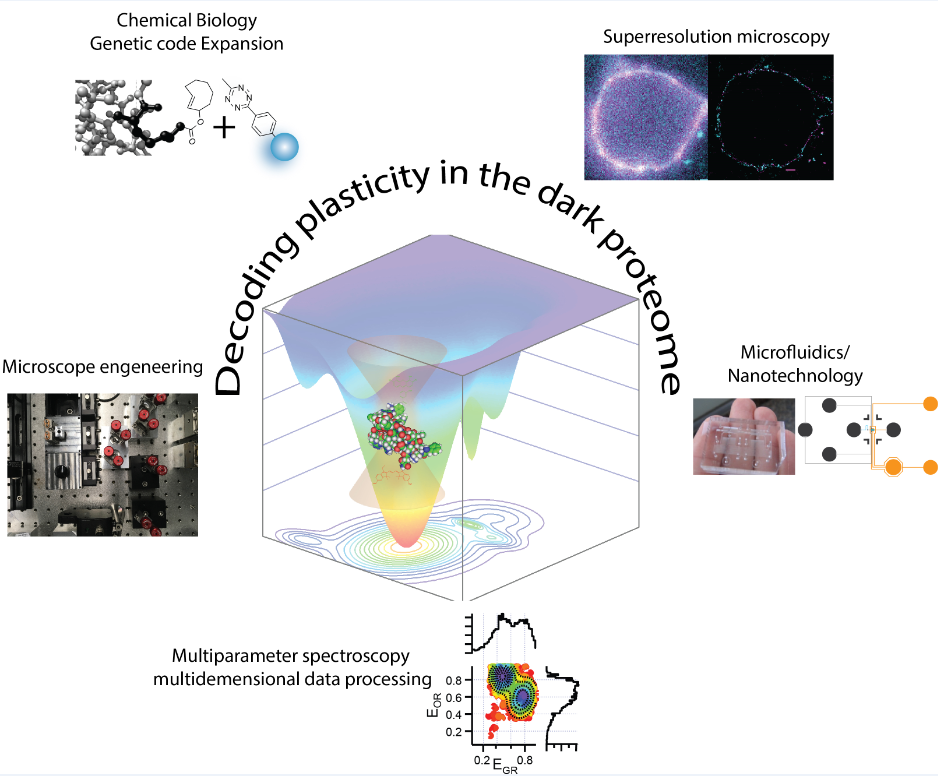
Our precision tools enable us to make even the most complex molecular machinery visible to our core methodologies, which are based on time-resolved multiparameter and nanoscopy tools. This enables innovative approaches to study the heterogeneity of IDPs in biology. More recently, we discovered a distinct ultrafast protein-protein interaction mechanism that can explain how nuclear pore complexes (NPCs) can efficiently fulfill their central role in cellular logistics and how nuclear transport can be both fast and selective at the same time. We also determined how this function can coexist with other nuclear transport mechanisms that provide a platform for cargo undocking. These findings provided a leap forward in our understanding of how the ability of IDPs to maintain different functionalities through conformational changes despite the normal requirement for rigid molecular specificity.
Despite our advancing technologies for in situ science, we always consider it important to perform studies on reconstituted systems (in vitro/biochemical) to understand our biological problems in a bottom-up fashion and complement our results from our in situ studies. To achieve this, we worked on various aspects e.g. i) utilizing microfluidics to integrate Lab-on-chip technology into our workflows and ii) developing a versatile baculovirus-based platform, which combines the benefits from GCE technology and the versatility of click chemistry with the strength of recombinant protein engineering. This technology now enables in vitro/bottom up/reconstitution based biological project design that was previously unachievable in situ. Our work is accompanied by rigorous analysis on the physicochemical properties of IDPs and examines to what extent simple known polymer concepts, such as phase separation, can be used to describe the function of such biopolymers in vivo.
Future Directions
IDPs are highly multifunctional. Due to their multivalency and large disordered regions they can function as dynamic scaffold platforms. We combine chemical and synthetic biology approaches to enable non-invasive, multi-color high- and super-resolution studies of activity-dependent changes of protein conformation in living cells, enabling fluorescence driven in situ Structural Biology. We will design new organelles so that we can control, tune and study the molecular mechanism of cellular function in a bottom up fashion. Ultimately the enhanced spatial and temporal resolution offered by our fluorescent methods will enable us to detect rare events and unexpected behaviours inside cells. We want to use this to identify and understand IDP multifunctionalities that are clearly distinguishable from their normal mode of action, as for example nucleoporins (Nups) in the nuclear pore complex (NPC). In fact, many IDP-Nups have roles in pathogen-host interactions and have been suggested to shuttle away from the NPC to function in gene regulatory processes. A prominent example of IDP-Nup action distinct from its normal mode in the NPC is Nup98, since several genetic fusions of Nup98 with transcription factors are related to leukaemia.
